Table Of Contents
An overview of PPRD
Database construction
Search single or multiple libraries
Search single or multiple genes
Search genes in selected libraries
Search genes in selected projects
Filter
IGV visualization
Data collection
Data analysis
Data analysis
References
Contact us
An overview of PPRD
Welcome to Arabidopsis RNA-Seq Database(PPRD), an online database for exploring 45,000+ published plant RNA-Seq libraries. PPRD is available at http://ipf.sustc.edu.cn/pub/plantrna/
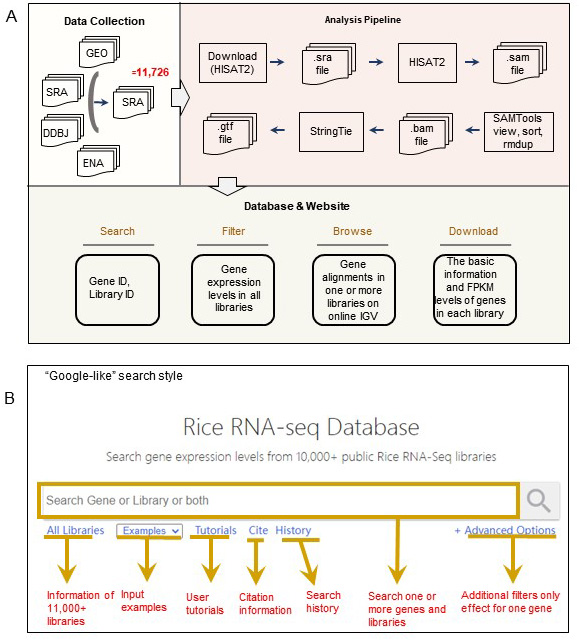
PPRD is a free, web-accessible, and user-friendly database, contributes to searching, filtering, visualizing, browsing, and downloading the RNA-seq data, is described in Figure 1A. PPRD functions are described as follows:
● PPRD consists of a large number of RNA-seq libraries of maize (19,664), rice (11,726), and soybean (4,085), wheat (5,816), and cotton (3,483).
● PPRD use a standardized pipeline to calculated the expression level of each gene in each library.
● PPRD can search the input gene ID and library ID, and return the basic description and expression levels.
● PPRD supports to download and share the search results.
● PPRD also has a built-in IGV-web interface.
● PPRD also supports conveniently accessing its homologs in other species to view its expression.
Database construction
PPRD contributes to search, visualize, browse, and download gene FPKM data, as Figure1A described. PPRD operates flexible that supports a "Google-like" search through querying of genes and libraries (Figure 1B), and returns the fundamental description, exhibit and visualize expression levels, and support IGV-web (https://igvteam.github.io/igv-webapp/) interfaced genomic alignment. We show the overflow in Figure2.
Search single or multiple libraries
PPRD also supports the search for one or more libraries. The input is one or more libraries ID separated by commas or spaces. All genes FPKM will be shown (see Figure 2) after the user submitted the library ID. By the way, gene and library ID can be queried together, and Figure 2B has demonstrated the result.

Search single or multiple gene
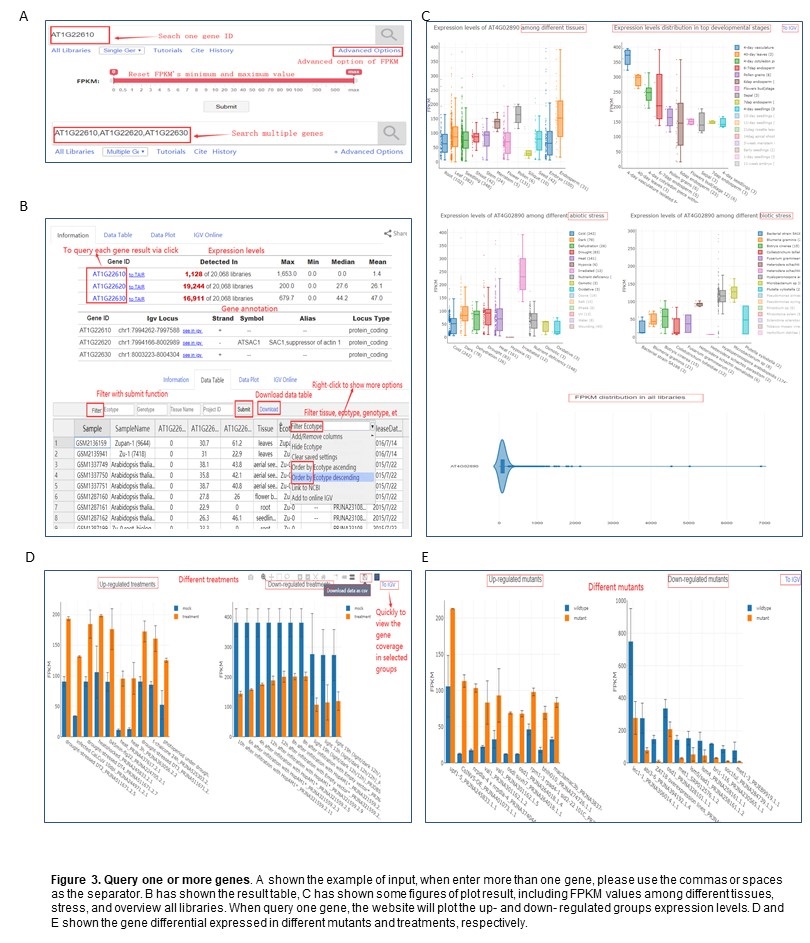
PPRD allows searching one or more genes annotated (Figure 3A). For visualization result, scatter and box plots describe the global FPKM levels (Figure 3C).
When only one gene has been searched, the advanced option will be valid to specify the maximum and minimum values of FPKM in the table.

Search genes in selected libraries
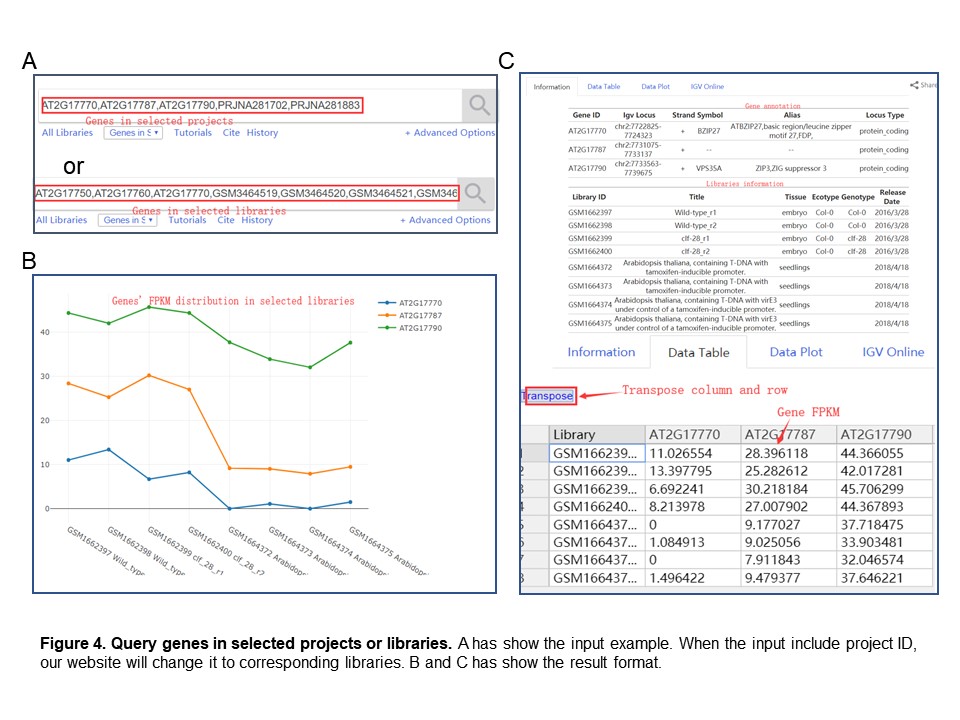
In PPRD, user can enter one or more genes with specify libraries, and that command must be separated by commas or spaces. Figure 4 has show the input example and result.
Search genes in selected projects
Our database also supports searching for the expression of single or multiple genes in a given project list. The input need to be separated by commas or spaces. Its result format is consistent with "Search genes in selected libraries", as shown in the Figure 4.
Filter
In the data table result, you can right-click to show advanced options for each column, such as hide, remove, order-by, filter. For the filter option, the user can select which options to view by selecting that from their respective drop-down boxes (Figure 3B).
IGV visualization
PPRD integrates an online IGV interface to browse and compare the genome matched RNA-Seq in one library or project. The links of online IGV interface are added into the results of each type of query. If user wants to clear all current tracks, please clicking the "clear tracks" button. Also, the IGV online can add multiple libraries or projects convenient for comparison(Figure 2D).
Data collection
We collected the five plants RNA-Seq datasets published till December 2020 from GEO, DDBJ, EBI, and SRA database using below keywords for each plant. After the search, we checked the detailed information and removed pseudo libraries, such as small RNA-seq or ncRNA-seq.
For rice RNA-seq: ((rice[Organism]) AND transcriptomic[Source]) AND rna seq[Strategy];
For soybean RNA-seq: ((Glycine[Organism]) AND transcriptomic[Source]) AND rna seq[Strategy];
For maize RNA-seq: ((maize[Organism]) AND transcriptomic[Source]) AND rna seq[Strategy];
For cotton RNA-seq: ((cotton[Organism]) AND transcriptomic[Source]) AND rna seq[Strategy];
For wheat RNA-seq: ((wheatOrganism]) AND transcriptomic[Source]) AND rna seq[Strategy];
Data analysis
Because the HISat2 supports access SRA database and can use the .sra format as input, we don't need to download raw data and convert the .sra to fastq format extra. (Because of the internet limit, some local servers can't support that function, at that time, you must use wget or other software to download raw data.) First, the raw reads were aligned to the reference genome, including Nipponbare of rice (Os-Nipponbare reference IRGSP-1.0), B73 of maize (B73 v4), Williams82 of soybean (Wm82.a2.v1), Triticum aestivum of wheat (IWGSC.v51), or Gossypium hirsutum of cotton (UTX-TM1_v2.1). And then, the FPKM (Fragments Per Kilobase of transcript per Million mapped reads) of whole-transcriptome was calculated for maize (45,953 genes), rice (55,801 genes), soybean (55,589 genes),wheat (120,744 genes), and cotton (74,902 genes) by Stringtie, respectively. The flowchart of the data collection, processing, and database functions is illustrated in Figure 1.
Coexpression analysis:
The R package WGCNA (version 1.69) (Langfelder and Horvath, 2008) was utilized to perform weighted gene co-expression analysis. We filtered the libraries with low-quality and low coverage to reduce data noise as previously (Zhang et al., 2020). Then remaining libraries were used to construct co-expression network. In brief, we obtained the optimal soft threshold power for each species based on the scale-free topology criterion. Then, the function blockwiseModules in WGCNA package was used to construct the weighted gene co-expression network and identify the modules with following parameters "TOMType = "unsigned", networkType = "unsigned", maxBlockSize = 40,000, minModuleSize = 30" (Zhang et al., 2020).
References
Kim, D., Langmead, B., and Salzberg, S.L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357-360.
Pertea, M., Pertea, G.M., Antonescu, C.M., Chang, T.C., Mendell, J.T., and Salzberg, S.L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33, 290-295.
Robinson, J.T., Thorvaldsdottir, H., Wenger, A.M., Zehir, A., and Mesirov, J.P. (2017). Variant Review with the Integrative Genomics Viewer. Cancer Res 77, e31-e34.
Langfelder, P. and Horvath, S. (2008) WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 9, 559.
Zhang, H., Zhang, F., Yu, Y., Feng, L., Jia, J., Liu, B., et al. (2020) A comprehensive online database for exploring ∼20,000 public Arabidopsis RNA-Seq libraries. Mol Plant.
Contact us
Dear the PPRD users,
Thank you for using the PPRD database!
If you encounter any problem or have any question, please don't hesitate to contact us at
zhaijx@sustech.edu.cn or zhangh9@mail.sustech.edu.cn.
Best wishes,
PPRD Team