Table Of Contents
An overview of ASRD
Data collection
Data processing
Data analysis
Database construction
Search sRNA and miRNA
Search library
Search gene
IGV online interface
References
Contact us
Please cite
Tutorials for ASRD users
An overview of ASRD
Welcome to the Arabidopsis Small RNA Database (ASRD), an online database with integrated, multi-faceted functions for exploring 2,000+ published Arabidopsis small RNA sequencing (sRNA-seq) libraries. ASRD is available at http://ipf.sustech.edu.cn/pub/asrd.
ASRD is a free, web-accessible, and user-friendly database that mainly contains three parts, including data collection, data processing, and database construction, and contributes to searching, filtering, browsing, visualizing, and downloading the sRNA-seq data (Figure 1). ASRD functions are described as follows.
• ASRD integrates 2,357,941,025 genome-matched sRNAs representing 254,678,199 unique sRNAs in 2,024 Arabidopsis sRNA-seq libraries. • ASRD uses a unified pipeline to process and analyze raw sRNA-seq data so that the normalized abundances of sRNAs can be compared across all libraries. • ASRD supports a "Google-like" search through querying of a single sRNA sequence, miRNA ID, miRNA name, miRNA sequence, gene ID, library ID, or library-related keyword. • ASRD also supports "Advanced options" for narrowing down the search area. • ASRD has abilities to display, visualize, and download the sRNA-seq data and search results. • ASRD integrated an online Integrative Genomics Viewer (IGV) browser for the convenience of browsing genome-matched sRNAs.

Figure 1. Overflow of the Arabidopsis small RNA Database (ASRD). A total of 2,024 publicly available Arabidopsis sRNA-seq libraries were collected from GEO and SRA databases, and processed with a unified pipeline for cross-library comparisons, and all the sRNA related information can be accessed via keyword-based searching on the ASRD website (http://ipf.sustech.edu.cn/pub/asrd).
Data collection
We collected Arabidopsis sRNA-seq data published until July, 2019 from GEO and SRA databases by searching with the following combinations of keywords "((sRNA) OR (sRNAs) OR siRNA OR smallRNA OR smallRNAs OR miRNA OR sRNA OR sRNAs OR siRNAs OR miRNAs) and Arabidopsis" (Figure 1). We obtained a total of 2,024 non-redundant libraries from Illumina NGS platform with raw sequencing data. Because the GEO and SRA databases share some libraries, we gave priority to name libraries with the GEO accession number.
Data processing
The raw sRNA-seq data in sra format were downloaded, processed, and analyzed by in-house scripts (all of our scripts can be shared upon request). Figure 1 describes the pipeline of sRNA-seq data processing. In brief, we used the fastq-dump of SRA Toolkit (2.8.2; https://www.ncbi.nlm.nih.gov/books/NBK158900) to convert raw data from sra to fastq format; if 3' adapter sequence hasn't been provided, we would predict and trim it with DNApi (Tsuji and Weng, 2016) and Cutadapt v1.16 (Martin, 2011), respectively, and chopped the 5' barcode if there exists. We then processed the remaining 18 to 28 nt reads in the fasta file to tag_count format. Then we mapped these reads to the Arabidopsis reference genome (TAIR10) using Bowtie v1.2.1.1 (Langmead et al., 2009), allowing zero mismatches (-v 0) and multiple hits (-a). We used Araport11 annotated t/r/sn/snoRNAs to flag corresponding types of sRNAs in each bam file. Finally, our database contains 2,357,941,025 genome-matched sRNAs representing 254,678,199 distinct sRNAs.
Data analysis
The annotation of 426 mature Arabidopsis miRNAs, including both miRNA and miRNA* sequences (named as miRNA-5p and miRNA-3p), was obtained from miRbase (version 22.1)(Kozomara et al., 2019), eight TAS loci: TAS1a, TAS1b, TAS1c, TAS2, TAS3, TAS3b, TAS3c, and TAS4 were used for calculating ta-siRNA abundance. The annotations of 38,621 genes were from Araport11. The list of 7,632 P4-siRNA loci was the same as previously described (Zhai et al., 2015). The 27,655 protein-coding (PC) genes and 3,901 transposon element genes (TEs) annotated in Araport11 were used to calculate the abundance of PC gene-generating siRNAs (PC-siRNAs) and TE-generating siRNAs (TE-siRNAs), respectively. The abundance of sRNAs in each library was calculated as TPM by normalizing to the total number of genome-matched reads excluding t/r/sn/snoRNA-derived ones. The TPM of sRNAs at a given locus was the sum of genome hits-normalized TPMs for all mapped reads at that locus.
Database construction
ASRD construction includes searching, filtering, browsing, visualizing, and downloading the sRNA-seq data of 2,024 libraries (Figure 1). The detailed functions of ASDR are described in Figure 2. ASRD supports a "Google-like" search through querying of a single sRNA sequence, miRNA ID, miRNA name, miRNA sequence, gene ID, library ID, or library-related keyword. The various types of queries are described as follows.
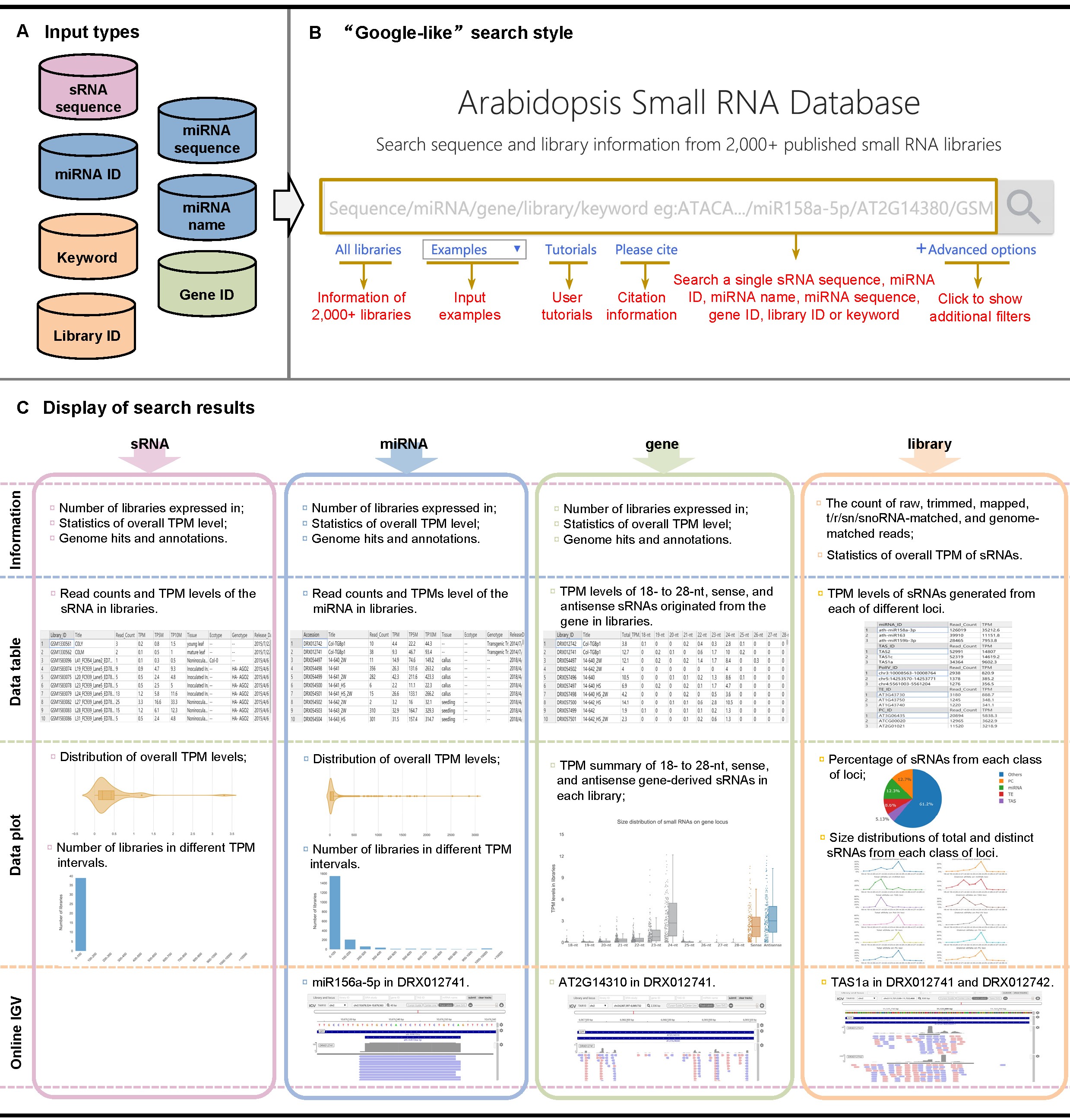
Figure 2. Basic functions of ASRD. A.Input can be a single sRNA sequence, miRNA ID, miRNA name, miRNA sequence, gen/e ID, library ID, or keyword. B. The introduction of ASRD main page functions. ASRD supports a "Google-like" search that allows the users to search each item in one single input box. "All libraries" shows the detailed information of libraries; "Examples" presents different types of queries; "Tutorials" links to instructions to users; "Advanced options" describes various additional filters. C. Display four types of search results, including the information, data table, data plot, and online IGV, based on different queries of sRNA, miRNA, gene, and library. All results can be downloaded to a local computer.
Search sRNA and miRNA
For querying the information of a single miRNA or sRNA, ASRD supports searches by a single sRNA sequence, miRNA ID, miRNA name, or miRNA sequence, and it will return the statistics of expression levels across all libraries. Taking the query of miR158a-5p as an example, Figure 3A shows the maximum, median, mean, and minimum TPM levels of miR158a-5p in all libraries. The violin plot shows the overall TPM distribution of miR158a-5p, and the bar diagram displays the number of miR158a-5p expressed libraries grouped by different TPM intervals (Figure 3B). The result table with the raw count, TPM, TP5M, and TP10M of miR158a-5p in each library is both online-available and downloadable. Also, the users can narrow down the query results via "Advanced options" by the combination of multiple filters of tissue, ecotype, genotype, release date, TPM level, and keyword (Figure 3C). Besides, the integrated online IGV interface can be used to browse the mapped miR158a-5p sequences on the genome (Figure 3D).
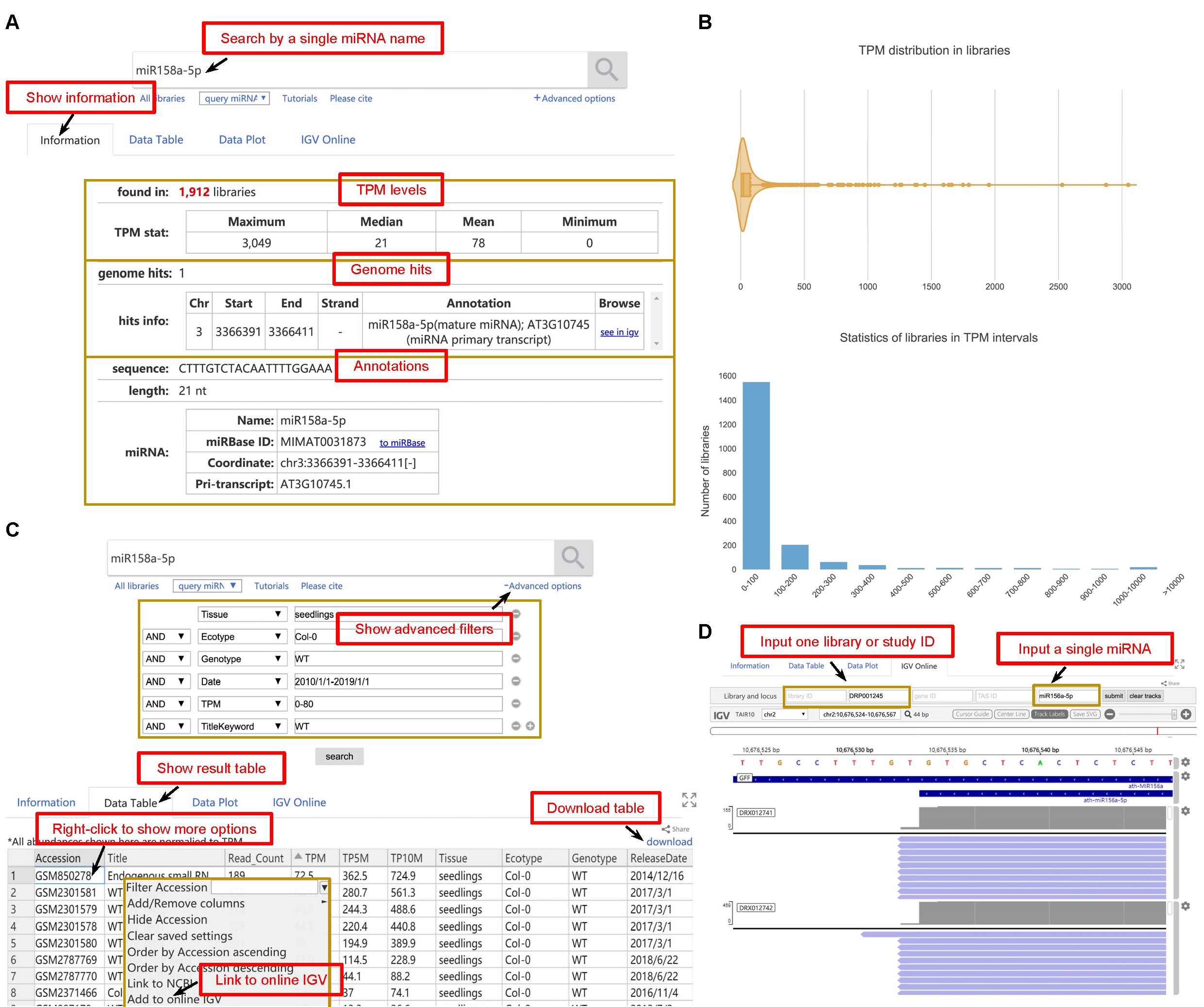
Figure 3. Example of a query using miR158a-5p. A. Example data for miR158a-5p, such as the statistics of maximum, median, mean, and minimum TPM levels in all libraries, as well as genome hits and its annotations on miRBase. B. The violin plot shows the overall TPM distribution, and each point represents the TPM level of miR158a-5p in a library. The bar diagram displays the number of libraries in different TPM intervals. C. The result table displays the abundance levels of miR158a-5p with adjusted different sequencing depth, including read count, TPM, TP5M, and TP10M. The advanced options can be used to filter the results by tissue, ecotype, genotype, release date, TPM level, or keyword. Right-clicking on each column of this table shows more operations, such as adding, removing, or sorting a column, linking a library to NCBI, or adding a library to online IGV. D. The online IGV browses the mapped miR158a-5p sequences in DRX012741 and DRX012742 libraries.
Search library
This function supports querying one library ID or library-related keyword. For one library, ASRD shows not only the library-related information gathered from the GEO or SRA database but the statistics of sRNA expression levels. Read counts include the raw, trimmed, mapped, t/r/sn/snoRNA-matched, genome-matched total and distinct reads, and TPM levels are for sRNAs generated from miRNA, PC, Pol IV, TE, ta-siRNA-generating loci (TAS), and others. The files of raw, trimmed, and mapped reads can be easily localized and fed into other tools that the users prefer (Figure 4A). For characterizing different classes of sRNAs, the pie diagram shows their percentage, and the line diagrams describe the size distributions of genome-matched total and distinct sRNAs For characterizing different classes of sRNAs, the pie diagram shows their percentage, and the line diagrams describe the size distributions of genome-matched total and distinct sRNAs (Figure 4B). ASRD also provides the read counts and TPM levels of sRNAs from each of miRNAs, TAS, and the Top-100 PC, Pol IV, and TE loci that ranked by TPM levels (Figure 4C).

Figure 4. Examples of queries using library ID and gene IDA-C. Search by one library. A-C. Search by library ID. A. The library information includes statistics of the raw, trimmed, mapped, t/r/sn/snoRNA-matched, genome-matched total and distinct reads, and the TPM levels of sRNAs generated from miRNA, PC, Pol IV, TAS, TE, and other classes of loci. The links to GEO or SRA official website and IGV online interface are shown here. B. The pie diagram exhibits the percentage of each class of sRNAs in the library, and the line diagrams show the size distributions of genome-matched total and distinct sRNAs from each class, respectively, with x-axis indicating sRNA size (nt), and y-axis showing the percentage of sRNAs. C. The tables display the read counts and TPM levels of sRNAs derived from each of miRNAs, TAS, and the Top-100 abundant PC, Pol IV, and TE loci. The advanced options additionally support the users to filter the search results by the TPM levels of sRNAs produced from different classes of loci. D-E. Search by gene ID.D. The diagram with scatters and boxes describes the TPM levels of sRNAs on the queried locus across all libraries. The grey, yellow, and blue color represents 18 to 28 nt sRNAs, sense, and antisense sRNAs, respectively. E. The table shows the TPM levels of 18 to 28 nt, sense, and antisense sRNAs generated from that locus in each library.
Search gene
ASRD supports searching by the IDs of genes annotated in Araport11 (Cheng et al., 2017). To explore sRNAs generated from a queried gene locus, the plot with scatters and boxes describes the TPM levels of 18 to 28 nt sRNAs across all libraries (Figure 4D), and the table detailed exhibits the size distribution of 18 to 28 nt, sense, and antisense sRNAs in each library. The same as a single miRNA query, this table also supports advanced filter and download (Figure 4E).
IGV online interface
ASRD integrates an online IGV interface to browse and compare the genome-matched sRNAs in one or more libraries. The users can find the link of an online IGV interface in the results of each type of query. It supports the browse of sRNAs on a given genomic region. For easy operation, we also extended IGV functions through submitting tracks of the library, SRA study, gene, and TAS ID, as well as miRNA name, and clearing all tracks that have been submitted. The SRA study track can be used for the users to add all libraries in a project at once when comparing the abundance of sRNAs generated from libraries belonging to the same project (Figure 5).

Figure 5. Online IGV functions. The library/study ID and gene ID/TAS ID/miRNA name support to be submitted individually or simultaneously. The browse result can be saved as a figure in SVG format by clicking at the button "Save SVG".
References
Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207-221 Axtell MJ (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64: 137-159 Axtell MJ, Jan C, Rajagopalan R, Bartel DP (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565-577 Baulcombe D (2004) RNA silencing in plants. Nature 431: 356-363 Bologna NG, Voinnet O (2014) The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol 65: 473-503 Borges F, Martienssen RA (2015) The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16: 727-741 Carthew RW, Sontheimer EJ (2009) Origins and Mechanisms of miRNAs and siRNAs. Cell 136: 642-655 Chen X (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25: 21-44 Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD (2017) Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89: 789-804 Clough E, Barrett T (2016) The Gene Expression Omnibus Database. Methods Mol Biol 1418: 93-110 Cuerda-Gil D, Slotkin RK (2016) Non-canonical RNA-directed DNA methylation. Nat Plants 2: 16163 Fei Q, Xia R, Meyers BC (2013) Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25: 2400-2415 Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Res 12: 996-1006 Kozomara A, Birgaoanu M, Griffiths-Jones S (2019) miRBase: from microRNA sequences to function. Nucleic Acids Res 47: D155-D162 Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database C (2011) The sequence read archive. Nucleic Acids Res 39: D19-21 Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523-536 Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 17: 3 Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394-408 Meyers BC, Axtell MJ (2019) MicroRNAs in Plants: Key Findings from the Early Years. Plant Cell 31: 1206-1207 Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC (2006) Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res 34: D731-735 Nelson ADL, Haug-Baltzell AK, Davey S, Gregory BD, Lyons E (2018) EPIC-CoGe: managing and analyzing genomic data. Bioinformatics 34: 2651-2653 Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29: 24-26 Ruiz-Ferrer V, Voinnet O (2009) Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol 60: 485-510 Tsuji J, Weng Z (2016) DNApi: A De Novo Adapter Prediction Algorithm for Small RNA Sequencing Data. PLoS One 11: e0164228 Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669-687 Zhai J, Bischof S, Wang H, Feng S, Lee TF, Teng C, Chen X, Park SY, Liu L, Gallego-Bartolome J, Liu W, Henderson IR, Meyers BC, Ausin I, Jacobsen SE (2015) A One Precursor One siRNA Model for Pol IV-Dependent siRNA Biogenesis. Cell 163: 445-455
Contact us
Dear the ASRD users, Thank you for using the ASRD database! If you encounter any problem or have any question, please don't hesitate to contact us at zhaijx@sustech.edu.cn or 11749333@mail.sustech.edu.cn. Best wishes, ASRD Team
Please cite
Feng L, Zhang F, Zhang H, Zhao Y, Meyers BC, Zhai J. An Online Database for Exploring Over 2,000 Arabidopsis Small RNA Libraries. Plant Physiol. 2020;182(2):685?691. doi:10.1104/pp.19.00959